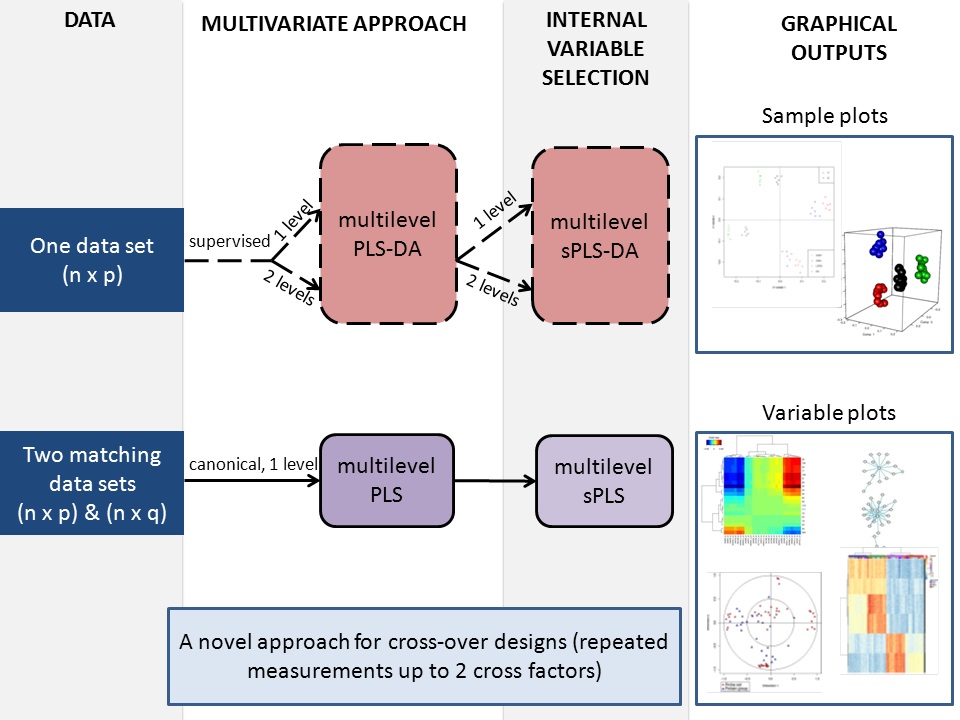
In order to take into account the complex structure of repeated measurements from different assays where different treatments are applied on the same subjects, a multilevel multivariate approach was developed to highlight the treatment effects within subject separately from the biological variation between subject (collaboration with A/Prof. B. Liquet).
Two different frameworks are proposed:
- a discriminant analysis (method = ‘splsda’) enables the selection of features separating the different treatments (indicated by the vector or the matrix ‘cond‘), whereas
- a integrative analysis (method = ‘spls’) enables the interaction of two matched data sets and the selection of subset of correlated variables (positively or negatively) across the samples. The approach is unsupervised: no prior knowledge about the samples groups is included.
The multilevel function first decomposes the variance in the data sets X (and Y) and applies either sPLS-DA or sPLS on the within-subject deviation. One or two-factor analyses are available for sPLS-DA.